Abstract
AIMS Antibiotic resistance is a global public health threat. There is sufficient evidence demonstrating the cause-effect relationship between antibiotic use and the development of hospital-acquired infections. However, it is unclear how the data on the relationship between antibiotic use, infection control practices and incidence of hospital-acquired infections can be used to inform daily clinical practice.
Method and results
Pubmed, Embase and the Cochrane Central Register of Controlled Trials (CENTRAL) databases, from inception until 30 May 2019 were searched. Studies with time-series analysis that have modelled the impact of hospital antibiotic use and infection control practices on the incidence of hospital-acquired resistant infections were considered and of 1327 records screened, 15 studies were eligible and included in this review.
Conclusion
The article outlines how the use of time series analysis can form the basis for a framework to develop strategies to intervene in order to tackle the problem of antibiotic use and resistance. Non-linear time-series analysis can provide a valuable tool to inform hospital antibiotic policies through identifying quantitative targets for optimising antibiotic use and controlling resistance.
Antibiotic resistance is a major threat to public health worldwide, causing increasing morbidity and mortality, and incurring significant healthcare costs. In a recent review aimed at estimating the global economic cost of antimicrobial resistance by 2050, the O’Neill report revealed that a continued rise in resistance would lead to 10 million deaths per year, resulting in a reduction of 2% in gross domestic product.1 In the UK, the Secretary for State for Health has launched new targets to reduce antimicrobial use in humans by 15% by 2024, and to halve the number of healthcare-associated gram negative bloodstream infections by 2025.2
The cause-effect relationships between antibiotic use and the development of healthcare-acquired infections (HCAIs) in hospitals has been widely evidenced.3–10 While it is clear that prudent antibiotic use and robust infection control practices are required, it is less clear how the data on relationships between antibiotic usage, infection control practices, and incidence of healthcare acquired infections can be used to inform best clinical practice. A key driver for informing the development of an efficient antibiotic policy, for example, should be the identification of the specific antibiotics that contribute most to the spread of local pathogen resistance, and following this strategies should be put in place to manage their use. In addition, healthcare systems are currently facing several challenges that would require policy makers to prioritise their interventions and provide efficient services. Antibiotic stewardship programmes were established to inform adequate antibiotic prescribing practices.8 The challenge for these programmes, however, is to balance providing adequate antibiotic treatment with preventing the emergence of antibiotic resistance.
Fortunately, mathematical models, including the analysis of time-series, have provided a greater understanding of population dynamics of pathogen transmission and the potential impact of specific interventions targeting these pathogens.7,11–14
The aim of this review is to summarise the published literature on the role of time-series analysis in the assessment of antibiotic use and resistance, in addition to highlighting relevant issues with antibiotic use, infection control and the development of HCAIs. A further aim is to demonstrate the utility of time-series analysis to provide interventions necessary for the management of HCAIs in hospitals.
Methods
Pubmed, Embase and the Cochrane Central Register of Controlled Trials (CENTRAL) databases, from inception until 30 May 2019 were searched (see Appendix 1 for full search strategy). Cohort studies with time-series analysis that have modelled the impact of hospital antibiotic use and infection control practices on the incidence of hospital-acquired resistant infections were included. Studies were excluded if: 1) they were published in a language other than English; 2) have used an interrupted time-series analysis; and 3) have investigated patients with community acquired resistant infections. Two authors (HA and FJ) independently screened the titles and abstracts of search results using the Rayyan website for systematic reviews,15 and then full texts of potentially eligible studies. A third reviewer (MA) resolved any disagreement. Data was synthesised narratively and based on the extracted results, textual summaries and tables were developed.
Results
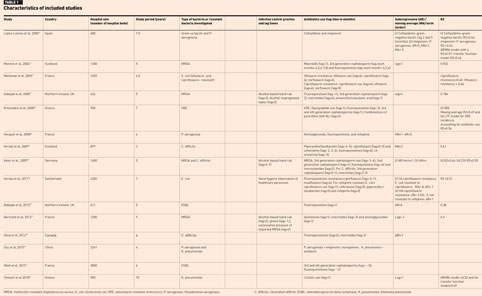
A total of 1327 records were identified from the initial database search. A total of 954 remained after duplicates were removed and were screened by titles and abstracts. After screening, 924 records were excluded, and 30 records were assessed for eligibility and after excluding a further 15 articles, 15 eligible articles remained and were included in the narrative analysis (Figure 1).
Studies reported the number of hospital beds (hospital size) with the minimum hospital size being 400 hospital beds16 and the maximum being 3000 hospital beds.17 The included studies were carried out mainly in hospitals in European countries (13 out of 15 studies) with, one study in Canada and one in China. The mean study periods in years (min–max) was 6 years (3–15 years). All studies reported the type of bacteria investigated, with several studies investigating more than one organism:
- Four studies investigated methicillin-resistant Staphylococcus aureus (MRSA)18–21
- Three studies investigated Pseudomonas aeruginosa16,22,23
- Three studies investigated Clostridium difficile20,24,25
- Two studies investigated Klebsiella pneumonia23,26
- Two studies investigated Escherichia coli27,28
- Two studies investigated extended spectrum beta lactamase (ESBL) producing bacteria17,29
- One study investigated gram negative bacilli16
- One study investigated vancomycin-resistant enterococci.30
Of the included studies, four studies19–21,28 reported the type of infection control practice implemented in the hospital. All studies16–30 used a linear time-series analysis and were conducted in one of the Organisation for Economic Cooperation and Development countries (OECD) (Table 1).
The included studies assessed different kinds of antibiotics used in the hospitals. Mainly, the studies assessed fluoroquinolones, 3rd and 4th generation cephalosporins, combinations of antibiotics (for example, amoxicillin and clavulanic acid or piperacillin and tazobactam), and macrolides (Table 1).
Discussion
This literature review summarises the work that has been done on the analysis of linear time-series. Several important themes are highlighted in this discussion.
Modelling antibiotic use, infection control and resistance
The criteria for the causal nature of an association were summarised as follows:
- coherence with existing information available in the literature or biological plausibility
- consistency of the association: results should be able to be reproduced in different settings, using different study designs and in different populations, (iii) time sequence: the cause must precede the effect
- specificity: refers to the question of whether the cause ever occurs without the presumed effect and whether the effect ever occurs without the presumed cause, and
- strength of the association, which included three concepts: its quantitative strength (that is, the effect size), dose–response (that is, an increase in the intensity of an exposure results in an increased risk of the effect under study) and study design (that is, controlling for bias and confounding factors, and taking account of the study design which was used to answer the research question).31 It is of note that not all the criteria have to be met to establish causality, but equally, meeting only one criterion is not sufficient to establish causality; the more criteria that are met, the more likely that the association is a causal one.31
Proving a causal relationship between antibiotic use and the development of resistance and HCAIs is difficult, as temporally sequenced observations on HCAIs are dependent on a number of factors. The incidence of Clostridium difficile infections (CDI) in December in any one year may have an effect on the incidence of CDI several months later. This is an effect known as autocorrelation and as classical statistical techniques rely on independent observations, they cannot be used to analyse such data.3,7,11,14,16,29 Over several years, time series analysis has been proposed by López-Lozano and colleagues as a suitable method to investigate the relationship between antimicrobial use and resistance.16,32 A time series is a collection of observations made sequentially over time, which are taken at equally spaced intervals much shorter than the overall study period.33 This type of analysis relates to a group of techniques aimed at adjusting a mathematical model to a time series, with the purpose of predicting the future behaviour of the series based on its historical behaviour, and trying to explain the characteristics and factors influencing the series.16
Application of time series analysis to antibiotic use and resistance
Epidemiological data are often generated in the form of time series, for example, notifications of disease, entries to a hospital, mortality rates etc. and are frequently collected at regular intervals, for example, weekly, monthly, yearly.34 Data produced by surveillance activity (for example, incidence rates of pathogens, antibiotic defined daily doses), which represent repeated measurements at regular intervals for specific periods, fits the definition of time series mentioned earlier.34 The effect of antimicrobial use on resistance is not simultaneous as an antimicrobial needs time to modify the ecological distribution of strains in a particular setting. The latter involves administration of an antibiotic that will cause modification of a patient’s bacterial population, which may result in the acquisition of resistance to the recently administered agent. A period of time is then necessary to pass the resistant organism from the patient into the surrounding environment where the environmental reservoir will accumulate, or alternatively, the organism may be passed from one patient to another via the hands of healthcare workers.35–37 Consequently, a delay may be observed between administration of an antibiotic and the spread of resistance.32 The same is true for the relationship between antibiotic use and the development of CDI. Environmental contamination, patient colonisation, exposure to antibiotics, symptomatic infection and diagnosis have to occur. All of these steps can contribute to the delay between antibiotic use and the resultant identification of CDI infection.24
In 1976, Box and Jenkins provided a practical method for modelling time series using Autoregressive Integrated Moving Average (ARIMA) models which are also called Box-Jenkins models.38 Since then, time series analysis has been applied in different fields such as industry and economics.34 There has also been increasing interest in using time series analysis in medical specialties such as endocrinology, cardiology and environmental medicine.39,40 In addition, it is possible to assess the relationships between a response series (outcome) and several explanatory (input) series by using transfer function analysis.39 Haugh proposed a practical method to build a transfer function model which has been applied in medicine to assess, for example, the relationship between influenza virus and mortality.41,42 The strategy for constructing ARIMA models requires three steps: (i) the identification of a particular model of the ARIMA class from the observed time series; (ii) the estimation of the parameters of the identified model; and (iii), diagnostic checks in order to determine whether or not the chosen model adequately represents the given set of data.16 This type of analysis requires 50 or more observations or time intervals in the series.43
Time-series analysis using ARIMA modelling can assess the lagged relationship between two variables. It therefore, enables measurement of a non-contemporaneous relationship between two or more variables measured on a temporal basis at the same frequency.32 ARIMA models allow the assessment of the behaviour of a series based on its historical characteristics and the present/past evolution of one or more predictor series. In 2000, López-Lozano and colleagues published the results of an application of time-series analysis to antibiotic resistance and antibiotic use data.16 In this research, the authors studied the relationship between hospital ceftazidime use and the percentage of ceftazidime-resistant gram-negative bacilli. In order to study the relationship between the two series, an ARIMA model was built and this model showed a significant cross-correlation between the two series with a lag (that is, time required to observe an effect) of one month and a coefficient of 0.42. The authors concluded that an increase (or decrease) of one defined daily dose (DDD) of ceftazidime/1000 patient-days during the previous month resulted in a 0.42 increase (or decrease) in the percentage of ceftazidime-resistant gram-negative bacilli during the following month.16 In recent years, the use of ARIMA modelling has been applied across Europe to investigate the temporal impact of antibiotic use and infection control practices, on the development of HCAIs.3,20,24 Such work has demonstrated that data observed in one month is a function of data observed in the previous months (autocorrelation) with a certain lag time. Thus, addressing the autocorrelation and the lag time which exists between the consecutive observations was an important aspect in order to identify the exact relationships.
Since the Monnet et al investigation in 2004,18 this method has been developed further through the inclusion of proxy measures for infection control activity19,20,24 patient group characteristics (that is, comorbidity/age-adjusted comorbidity index)29 and the interaction between antibiotic prescribing in the community and levels of pathogens in hospitals.28,29 A retrospective quasi-experimental study was undertaken in the Northern Health and Social Care Trust in Northern Ireland in order to study the effect of antibiotic use and infection control practices on the incidence of hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA).19 This study involved the assessment of the impact of antibiotic use on the spread of MRSA while combining data on proxy measures for infection control practices (for example, alcohol-based hand rubs, glove use, disinfectant use, screening for MRSA activity). Following the application of a multivariate ARIMA modelling to these data, the authors were able to demonstrate the impact of several influential variables involved in the transmission of MRSA and presented the relative efficacy of different MRSA-controlling measures.
In addition, the authors of this research were able to provide a possible way forward for estimating the number of patients needed to be treated with specific antibiotics in order to cause the incidence of a pathogen case, within a specific timeframe.19 For example, their analysis showed that treatment of 30 patients with fluoroquinolones would result in the occurrence of one HA-MRSA case within an average of one month, whilst treatment of 67 patients with macrolides would result in the development of one HA-MRSA case, over an average time frame of four months. The latter estimation of size-effects, and required time frame to observe an effect, has important clinical implications as it will guide the decision of classifying the identified antibiotics into risk groups (high, medium, low).
In addition, measuring the delay required to observe an effect following the restriction/use of particular antibiotics would assist in determining the optimal time required for an antibiotic restriction policy. This study demonstrated the power and utility of using time series methods to assess the combined impact of infection control and antibiotic use on resistance rates, an area where demonstrating causality has proven difficult. Other studies were aimed at studying the role of infection control practices, combined with antibiotic use, on spread of resistant pathogens.20,24 Interestingly, a possible way for comparing the time-series analysis results between different healthcare settings has been suggested.44 This was achieved through the determination of the dose-response relationship between antibiotic consumption and the emergence of resistance.
A dose–response relationship can be defined as the percentage change in the probability of an organism being resistant following a 1% change in the level of antibiotic use.44
Recently, in work undertaken in Scotland, Lawes et al explored the relationship between antibiotic use and MRSA, with the authors proposing a logical extension of the time series analysis methods named as non-linear time-series analysis. The latter approach considers the fact that the nature of the relationships between antibiotic use and resistance, in complex natural systems, are likely to be non-linear. By using the non-linear time series analysis, it was possible to demonstrate that reducing antibiotic selection pressures below identified thresholds allowed the prediction of shifts in the molecular and clinical epidemiology of Staphylococcus aureus and Clostridium difficile. Importantly, the authors managed to create thresholds of antibiotic use (points at which a further reduction in antibiotic use has minimal impact on the development and spread of resistance) to control MRSA rates.13,45
In a landmark study, a non-linear time-series analysis approach for identifying thresholds using historical prescribing and microbiological data from five populations in Europe was undertaken. Minimum thresholds in temporal relationships between use of selected antibiotics and rates of carbapenem-resistant Acinetobacter baumannii (in Hungary), extended spectrum β-lactamase producing Escherichia coli (Spain), cefepime-resistant Escherichia coli (Spain), gentamicin-resistant Pseudomonas aeruginosa (France), and MRSA (Northern Ireland) in different epidemiological phases were identified.46
Conclusions
HCAIs are considered a major problem worldwide, with high clinical and financial burden on healthcare authorities. The analysis of time-series approach can form the basis for a framework to provide several strategies to intervene in order to tackle the problem of antibiotic use and resistance. By using routinely generated data, it is possible to identify minimum thresholds in the relationship between the use of selected antibiotics and rates of HCAIs in hospitals. By identifying a threshold in antibiotic use, hospital guidelines can be informed accordingly, aiming at removing the selection pressure of the identified antibiotic, thus, controlling rates of specific healthcare acquired infections in hospitals and improving patient outcomes. Non-linear time-series analysis can provide a valuable tool to inform hospital antibiotic policies through identifying quantitative targets
for optimising antibiotic use and controlling resistance.
Key points
- Hospital-acquired infections impose high clinical and financial burdens on healthcare authorities.
- Routinely generated data in the hospital could be used to identify thresholds in the relationship between the use of antibiotics and rates of nosocomial infections.
- Non-linear time-series analysis models derived from historical data are a potential method to estimate the outcome of antibiotic restriction plans.
- Time-series analysis could provide greater understanding of population dynamics of pathogen transmission and the potential impact of specific interventions targeting these pathogens.
- Using thresholds in antibiotic use to inform hospital guidelines may result in the removal of selection pressures of the identified antibiotic, controlling rates of specific healthcare acquired infections in hospitals and improving patient outcomes.
Funding
This work was supported by a grant from Public Health Agency/ Northern Ireland.
Financial and conflicts of interest disclosure
None declared.
Authors
Feras Jassim Jirjees BSc MPharm PhD College of Pharmacy, University of Sharjah, Sharjah, United Arab Emirates
Hala Jehad Al-Obaidi BSc MSc Pharmacy PhD School of Pharmacy and Pharmaceutical Science, Ulster University, Northern Ireland, UK
Muhammad Sartaj MBBS MPH DHSCM FFPH Public Health Agency, Belfast, Northern Ireland, UK
Geraldine Conlon-Bingham MPharm MSc PhD Pharmacy Department, Southern Health and Social Care Trust, Northern Ireland, UK
David Farren MB MCh BAO MSc MRCP FRFPath Department of Medical Microbiology, Southern Health and Social Care Trust, Northern Ireland, UK
Michael Scott BSc PhD Regional Medicines Optimisation Innovation Centre, Northern Ireland, UK
Ian Gould FRCPath FRCPE DSc Medical Microbiology Department, Aberdeen Royal Infirmary, Aberdeen, Scotland, UK
José-María López-Lozano MD PhD Research Group on Health Sciences Data Analysis, Universidad de Murcia, Spain
Mamoon Aldeyab BSc Pharmacy MSc Clinical Pharmacy MSc Epidemiology PhD Department of Pharmacy, University of Huddersfield, Huddersfield, UK
References
- O’Neill J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations 2014. 18 June 2019. https://amr-review.org/Publications.html (accessed May 2020).
- HM Government. Tackling antimicrobial resistance 2019–2024 – The UK’s five-year national action plan 2019. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data /file/773130/uk-amr-5-year-national-action-plan.pdf (accessed May 2020).
- Aldeyab MA et al. Quasiexperimental study of the effects of antibiotic use, gastric acid-suppressive agents, and infection control practices on the incidence of Clostridium difficile-associated diarrhea in hospitalized patients. Antimicrob Agents Chemother 2009;53(5):2082–8.
- Chang S et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 2003;348(14):1342–7.
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 2004;10(12 Suppl):S122–9.
- Lipsitch M, Samore MH. Antimicrobial use and antimicrobial resistance: a population perspective. Emerg Infect Dis 2002;8(4):347–54.
- Lopez-Lozano JM et al. A nonlinear time-series analysis approach to identify thresholds in associations between population antibiotic use and rates of resistance. Nature Microbiol 2019;4(7):1160–72.
- MacDougall C, Polk RE. Antimicrobial stewardship programs in health care systems. Clin Microbiol Rev 2005;18(4):638–56.
- McGowan JE Jr. Antimicrobial resistance in hospital organisms and its relation to antibiotic use. REv Infect Dis 1983;5(6):1033–48.
- Tsiodras S et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 2001;358(9277):207–8.
- Aldeyab MA et al. Modelling the impact of antibiotic use and infection control practices on the incidence of hospital-acquired methicillin-resistant Staphylococcus aureus: a time-series analysis. J Antimicrob Chemother 2008;62(3):593–600.
- Bonovas S et al. Mathematical models for the evaluation of antibiotic resistance in hospitals: a systematic review. Central Eur J Publ Health. 2003;11(4):229–37.
- Lawes T et al. Turning the tide or riding the waves? Impacts of antibiotic stewardship and infection control on MRSA strain dynamics in a Scottish region over 16 years: non-linear time series analysis. BMJ Open 2015;5(3):e006596.
- Shardell M et al. Statistical analysis and application of quasi experiments to antimicrobial resistance intervention studies. Clin Infect Dis 2007;45(7):901–7.
- Ouzzani M et al. Rayyan – a web and mobile app for systematic reviews. Syst Rev 2016;5(1):210.
- Lopez-Lozano JM et al. Modelling and forecasting antimicrobial resistance and its dynamic relationship to antimicrobial use: a time series analysis.Int J Antimicrob Agents 2000;14(1):21–31.
- Vibet MA et al. Systematic analysis of the relationship between antibiotic use and extended-spectrum beta-lactamase resistance in Enterobacteriaceae in a French hospital: a time series analysis. Eur J Clin Microbiol Infect Dis 2015;34(10):1957–63.
- Monnet DL et al. Antimicrobial drug use and methicillin-resistant Staphylococcus aureus, Aberdeen, 1996–2000. Emerg Infect Dis 2004;10(8):1432–41.
- Aldeyab MA et al. Modelling the impact of antibiotic use and infection control practices on the incidence of hospital-acquired methicillin-resistant Staphylococcus aureus: a time-series analysis. J Antimicrob Chemother 2008;62(3):593–600.
- Kaier K et al. Two time-series analyses of the impact of antibiotic consumption and alcohol-based hand disinfection on the incidences of nosocomial methicillin-resistant Staphylococcus aureus infection and Clostridium difficile infection. Infect Control Hosp Epidemiol 2009;30(4):346–53.
- Bertrand X et al. Temporal effects of infection control practices and the use of antibiotics on the incidence of MRSA. J Hosp Infect 2012;82(3):164–9.
- Hocquet D et al. Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2008;52(3):1173–5.
- Zou YM et al. Trends and correlation of antibacterial usage and bacterial resistance: time series analysis for antibacterial stewardship in a Chinese teaching hospital (2009-2013). Eur J Clin Microbiol Infect Dis 2015;34(4):795–803.
- Vernaz N et al. Temporal effects of antibiotic use and Clostridium difficile infections. J Antimicrob Chemother 2009;63(6):1272–5.
- Gilca R et al. Seasonal variations in Clostridium difficile infections are associated with influenza and respiratory syncytial virus activity independently of antibiotic prescriptions: a time series analysis in Quebec, Canada. Antimicrob Agents Chemother 2012;56(2):639–46.
- Tansarli GS et al. Colistin resistance in carbapenemase-producing Klebsiella pneumoniae bloodstream isolates: Evolution over 15 years and temporal association with colistin use by time series analysis. Int J Antimicrob Agents. 2018;52(3):397–403.
- Mahamat A et al. Evolution of fluoroquinolone resistance among Escherichia coli urinary tract isolates from a French university hospital: application of the dynamic regression model. Clin Microbiol Infect 2005;11(4):301–6.
- Vernaz N et al. Modelling the impact of antibiotic use on antibiotic-resistant Escherichia coli using population-based data from a large hospital and its surrounding community. J Antimicrob Chemother 2011;66(4):928–35.
- Aldeyab MA et al. The impact of antibiotic use on the incidence and resistance pattern of extended-spectrum beta-lactamase-producing bacteria in primary and secondary healthcare settings. Br J Clin Pharmacol 2012;74(1):171–9.
- Kritsotakis E et al. The dynamic relationship between antibiotic use and the incidence of vancomycin-resistant Enterococcus: time-series modelling of 7-year surveillance data in a tertiary-care hospital. Clin Microbiol Infect 2008;14(8):747–54.
- Strom BL. Study designs available for pharmacoepidemiologic studies. Textbook of Pharmacoepidemiology 2013:17–29.
- López-Lozano J-M et al. Applications of time-series analysis to antibiotic resistance and consumption data. Antibiotic Policies: Springer; 2005:447–63.
- Chatfield C. The analysis of time series: an introduction. Chapman and Hall/CRC; 2003.
- Helfenstein U. The use of transfer-function models, intervention analysis and related time-series methods in epidemiology. Int J Epidemiol 1991;20(3):808–15.
- Boyce JM et al. Environmental contamination due to methicillin-resistant Staphylococcus aureus: Possible infection control implications. Infect Cont Hosp Epidemiol 1997;18(9):622–7.
- Huang SS, Datta R, Platt R. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med 2006;166(18):1945–51.
- Faires MC et al. The identification and epidemiology of meticillin-resistant Staphylococcus aureus and Clostridium difficile in patient rooms and the ward environment. BMC Infect Dis 2013;13(1):342.
- Box G, Jenkins GM. Holden-Day Series in Time Series Analysis, Revised ed. Holden-Day San Francisco, CA, USA; 1976.
- Helfenstein U. Box-Jenkins modelling in medical research. Stat Methods Med Res 1996;5(1):3–22.
- Crabtree BF et al. The individual over time: time series applications in health care research. J Clin Epidemiol 1990;43(3):241–60.
- Haugh LD. Checking the independence of two covariance-stationary time series: a univariate residual cross-correlation approach. J Am Stat Assoc 1976;71(354):378–85.
- Stroup DF, Thacker SB, Herndon JL. Application of multiple time series analysis to the estimation of pneumonia and influenza mortality by age 1962–1983. Stats Med 1988;7(10):1045–59.
- England E. How interrupted time series analysis can evaluate guideline implementation. Pharm J 2005;275(7367):344–7.
- Phelps CE. Bug drug-resistance – Sometimes less is more. Med Care 1989;27(2):194–203.
- Lawes T et al. Effect of a national 4C antibiotic stewardship intervention on the clinical and molecular epidemiology of Clostridium difficile infections in a region of Scotland: a non-linear time-series analysis. Lancet Infect Dis 2017;17(2):194–206.
- López-Lozano J-M et al. A nonlinear time-series analysis approach to identify thresholds in associations between population antibiotic use and rates of resistance. Nat Microbiol 2019;4(7):1160.